myXCLASSMapFit
Fitting multiple data cubes to generate parameter maps
In contrast to the myXCLASSFit function, the myXCLASSMapFit function offers the possibility to fit multiple frequency ranges in multiple FITS data cubes from multiple telescopes simultaneously. It can be used with different optimization algorithms to optimize the input parameters defined in an molfit file to achieve a good description of the observational data. Details of the myXCLASSFit are described in Sect. “myXCLASSMapFit”.
The myXCLASSMapFit function requires at least the following three input files
molfit file, see Sect. “The molfit file”, to define which molecules are taken into account and the corresponding components,
observational xml, see Sect. “Observational xml file” , to control the import of the observational data,
algorithm xml file, see Sect. “Algorithm xml file” to specify the optimization algorithm.
In addition to these three input files the following input files are required for some fits as well,
the iso ratio file for describing relations between molecules and their isotopologues, see Sect. “The iso ratio file”,
a ds9 [1] region or masking file, see Sect. “myXCLASSMapFit”.
At the end of the whole fit procedure, the myXCLASSMapFit function
creates FITS images for each optimized parameter, where each pixel corresponds
to the value of the optimized parameter taken from the best fit for this pixel.
The name of each parameter FITS image consists of the phrase BestResult
followed by the name of the parameter, e.g. T_rot for the rotation
temperature etc, the name of the corresponding molecule, and finally by the
index of the related component. For example, the FITS file
BestResult___parameter__T_rot___molecule__CH3OH_v=0____component__1.fits,
describes the optimized excitation (rotation) temperature of the first component of molecule CH\(_3\)OH\(_{v=0}\) for each pixel of the selected fit region. In addition to the parameter maps, the myXCLASSMapFit function creates one FITS image, where each pixel corresponds to the \(\chi^2\) value of the best fit, i.e. the quality of the fit, for the corresponding pixel.
Furthermore, the myXCLASSMapFit function creates FITS cubes for each
used algorithm and fitted data cube, where each pixel contains the modeled
spectrum. The name of these FITS cubes ends with the expression
__model.out.fits. (If more than one fit algorithm was used, this
expression is extended by the respective name of the optimization
algorithm). If more than one frequency range was used for a given FITS
cube, the name of the corresponding FITS cube describing the synthetic
spectra also contains the frequency limits (in MHz), i.e.
name-of-FITS-file__MinFreq_-_MaxFreq__MHz__model.out.fits.
The FITS files, which end with .chi2.out.fits, describe the
\(\chi^2\) function for each pixel.
In addition, the myXCLASSMapFit function converts the following parameters
N_tot,EM_RRL,nHcolumn_cont_dust,nHcolumn,
to a log scale, i.e. these parameter values are converted automatically to their log10 values to get a better fit. At the end of the fitting process, the log10 values are converted back to the original linear values. So, the XCLASS log files contain the log10 values of these parameters, whereas the input and output molfit files contain the linear values, respectively.
Example call of the myXCLASSFit function:
>>> from xclass import task_myXCLASSMapFit
>>> import os
## get path of current directory
>>> LocalPath = os.getcwd() + "/"
# define path and name of molfit file
>>> MolfitsFileName = LocalPath + "files/my_map-molecules.molfit"
# define path and name of obs. data file
>>> ObsXMLFileName = LocalPath + "files/my_observation__map.xml"
# define path and name of algorithm xml file
>>> AlgorithmXMLFileName = LocalPath + "files/my_algorithm-settings.xml"
## use fast-fitting method?
>>> FastFitFlag = True
## use full-fitting method?
>>> FullFitFlag = False
# define path and name of region file
>>> regionFileName = LocalPath + "files/my_map-region.reg"
# define lower limit for intensity (pixel with max. intensity below given
# limit are ignored)
>>> Threshold = 0.0
# define number of iterations to smooth parameter maps
>>> ParamMapIterations = 1
# define parameter used for smoothing
>>> ParamSmoothMethodParam = 1.0
# define scipy method ("gaussian", "uniform", "median") used for parameter
# map smoothing
>>> ParamSmoothMethod = "uniform"
# define path of a directory containing FITS images describing parameter
# maps to update the parameter defined in the molfit file for each pixel
>>> ParameterMapDir = ""
# call myXCLASSMapFit function
>>> JobDir = task_myXCLASSMapFit.myXCLASSMapFitCore( \
MolfitsFileName = MolfitsFileName, \
ObsXMLFileName = ObsXMLFileName, \
FastFitFlag = FastFitFlag, \
FullFitFlag = FullFitFlag, \
AlgorithmXMLFileName = AlgorithmXMLFileName, \
clusterdef = clusterdef, \
regionFileName = regionFileName, \
Threshold = Threshold, \
ParameterMapDir = ParameterMapDir, \
ParamMapIterations = ParamMapIterations, \
ParamSmoothMethodParam = ParamSmoothMethodParam, \
ParamSmoothMethod = ParamSmoothMethod)
# start myXCLASSMapFit GUI
>>> task_myXCLASSMapFit.StartPlottingGUI(JobDir)
myXCLASSMapFit GUI
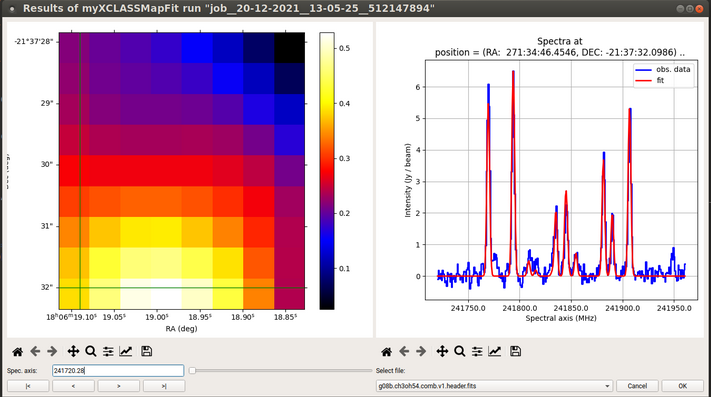
Fig. 7 Example of the myXCLASSMapFit-GUI.
In order to check the results of the fitting process, the myXCLASSMapFit function provides a graphical user interface to display the fitting results in conjunction with the observational data, see Fig. 1. By using the mouse on the left side, the user can select a specific pixel, whose spectrum together with the synthetic spectrum is then displayed on the right side. The parameter maps as well as the map describing the \(\chi^2\) values for all fitted pixels can be load into the left panel as well. When such a map is loaded into the left panel, the right panel still shows the spectra of the last imported FITS cube. The function only needs the path and the name of the job directory of the selected call of the myXCLASSMapFit function as input parameters.
myXCLASSRedoMapFit
This function offers the possibility to redo one or more so called
pixel fits of a previous myXCLASSMapFit run. The function performs
fits for the selected pixels and recreates the different parameter
maps using the new optimized parameter values together with the
corresponding FITS cube(s) describing the synthetic spectra for all
selected pixels. The names of these maps are created in the same way
as described in Sect. “myXCLASSMapFit”. In
addition, the myXCLASSMapRedoFit function renames the files
describing the parameter maps from the previous call of the
myXCLASSMapFit function by replacing the end of the each filename
".fits" by the phrase "__REDO-OLD.fits". The user has to
define the pixel coordinates, which should be re-fitted, the job
number of the previous myXCLASSMapFit run, and the new molfit
file.
Example call of the myXCLASSRedoMapFit function:
>>> from xclass import task_myXCLASSMapRedoFit
>>> from xclass import task_myXCLASSMapRedoFit
>>> import os
## get path of current directory
>>> LocalPath = os.getcwd() + "/"
# define path and name of molfit file
>>> MolfitsFileName = LocalPath + "files/my_map-molecules.molfit"
# define path and name of obs. data file
>>> ObsXMLFileName = LocalPath + "files/my_observation__map.xml"
# define path and name of algorithm xml file
>>> AlgorithmXMLFileName = LocalPath + "files/my_algorithm-settings.xml"
## use fast-fitting method?
>>> FastFitFlag = True
## use full-fitting method?
>>> FullFitFlag = False
# define path and name of region file
>>> regionFileName = LocalPath + "files/my_map-region.reg"
# define lower limit for intensity (pixel with max. intensity below given
# limit are ignored)
>>> Threshold = 0.0
# define path and name of so-called cluster file
>>> clusterdef = LocalPath + "files/clusterdef.txt"
# define number of iterations to smooth parameter maps
# (=1 means no smoothing)
>>> ParamMapIterations = 1
# define parameter used for smoothing
>>> ParamSmoothMethodParam = 1.0
# define scipy method ("gaussian", "uniform", "median") used for parameter
# map smoothing
>>> ParamSmoothMethod = "uniform"
# define path of a directory containing FITS images describing parameter
# maps to update the parameter defined in the molfit file for each pixel
>>> ParameterMapDir = ""
# call myXCLASSMapFit function
>>> JobDir = task_myXCLASSMapFit.myXCLASSMapFitCore( \
MolfitsFileName = MolfitsFileName, \
ObsXMLFileName = ObsXMLFileName, \
FastFitFlag = FastFitFlag, \
FullFitFlag = FullFitFlag, \
AlgorithmXMLFileName = AlgorithmXMLFileName, \
clusterdef = clusterdef, \
regionFileName = regionFileName, \
Threshold = Threshold, \
ParameterMapDir = ParameterMapDir, \
ParamMapIterations = ParamMapIterations, \
ParamSmoothMethodParam = ParamSmoothMethodParam, \
ParamSmoothMethod = ParamSmoothMethod)
# get job number for myXCLASSMapFit call
>>> JobDirName = JobDir.split("/")[-2]
>>> LocalJobNumber = JobDirName.split("__")
>>> LocalJobNumber = LocalJobNumber[-1]
# define the job number, e.g. the name of the job directory is
# "job__13-02-2020__15-16-12__444385099", with job number "444385099"
# Here, we use the job number from the previous myXCLASSMapFit call!
>>> JobNumber = LocalJobNumber
# define list of pixel which will be updated
>>> PixelList = [[1, 1], [0, 2]]
# define path and name of molfit file
>>> MolfitsFileName = LocalPath + "files/my_new-map-molecules.molfit"
# define max. number of iterations
>>> NumberIteration = 50
# set fast flag
>>> FastFitFlag = True
## call myXCLASSMapRedoFit function
>>> task_myXCLASSMapRedoFit.myXCLASSMapRedoFitCore( \
JobNumber = JobNumber, \
PixelList = PixelList, \
NumberIteration = NumberIteration, \
MolfitsFileName = MolfitsFileName, \
FastFitFlag = FastFitFlag)